Large language models (LLMs) adopted for specific enterprise applications most often benefit from model customization. Enterprises need to tailor LLMs for their specific needs and quickly deploy them for low-latency and high-throughput inferencing. This post will help you get started with this process.
Specifically, we’ll show how to customize the Llama 3 8B NIM for answering questions in the biomedical domain using the PubMedQA dataset. Question answering is crucial for organizations to quickly extract key information from large volumes of content and provide relevant information to customers.
NVIDIA software used in this tutorial
NVIDIA NIM, part of NVIDIA AI Enterprise, is a set of easy-to-use inference microservices designed to accelerate the deployment of performance-optimized generative AI models in enterprises. NIM inference microservices can be deployed anywhere, from workstations and on-premises to the cloud, providing enterprises with control over their deployment choices and ensuring data security. It also offers industry-leading latency and throughput, enabling cost-effective scaling and delivering a seamless experience for end users.
Today, users can access NIM inference microservices for the Llama 3 8B Instruct and Llama 3 70B Instruct models for self-hosted deployment on any NVIDIA-accelerated infrastructure. If you’re just getting started with prototyping, check out the Llama 3 APIs from the NVIDIA API catalog.
NVIDIA NeMo is an end-to-end platform for developing custom generative AI. NeMo includes tools for training, customization, retrieval-augmented generation (RAG), guardrails, toolkits, data curation, and model pretraining. NeMo provides an easy, cost-effective, and fast way to adopt generative AI.
Using NeMo framework, enterprises can build models that align with their brand voice and understand domain-specific knowledge. Whether creating a customer service chatbot or a bot for IT help, NeMo helps developers build custom generative AI that excels at its tasks while incorporating industry terminology, domain knowledge and skills, and unique organizational requirements.
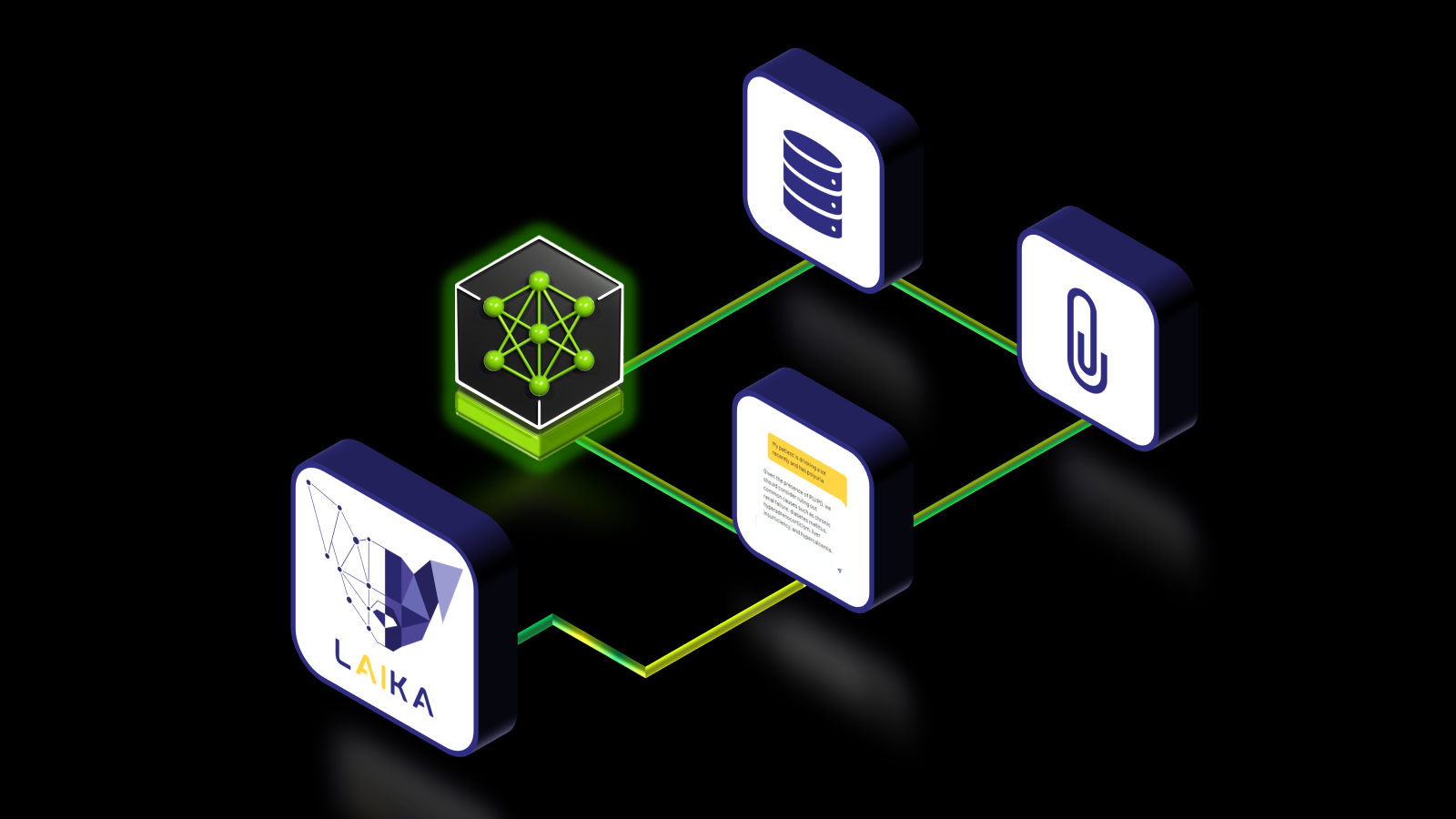
Figure 1 shows the general steps involved in customizing an LLM NIM with LoRA using NeMo and deploying it with NIM. At a high level, first convert the models to .nemo format. Then create LoRA adapters for NeMo models, and use these adapters with NIM for inference on the customized model. NIM supports dynamic loading of LoRA adapters, enabling training of multiple LoRA models for different use cases.

Prerequisites
Before you begin, ensure you have the following:
- Access to an NVIDIA A100, NVIDIA H100, or NVIDIA L40S GPU. It’s recommended to have cumulative memory of at least 80 GB across one or more GPUs.
- A Docker-enabled environment, with NVIDIA Container Runtime installed. This will make the container GPU-aware.
- An NGC CLI API key, provided when you authenticate with NVIDIA NGC and download the NGC CLI tool.
- An NVIDIA AI Enterprise license. To request a free 90-day trial license, visit Llama 3 8B Instruct on the API catalog and click the Run Anywhere with NIM button.
Step 1: Download the Llama 3 8B Instruct model
You can download the Llama 3 8B Instruct model from the NVIDIA NGC catalog using the CLI. It’s already converted to .nemo format, compatible with NeMo framework.
ngc registry model download-version "nvidia/nemo/llama-3-8b-instruct-nemo:1.0"
This will create a folder named llama-3-8b-instruct-nemo_v1.0, which includes the .nemo file.
Step 2: Get the NeMo framework container
NeMo framework is available as a Docker container on the NGC catalog. The environment and all the scripts for LoRA fine-tuning are included in this container.
The following code assumes that the Llama 3 8B Instruct model folder is part of the current working directory, so it’s mounted (in /workspace) and the fine-tuning scripts can access it.
# Run the docker container in interactive mode
docker run \
--gpus all \
--shm-size=2g \
--net=host \
--ulimit memlock=-1 \
--rm -it \
-v ${PWD}:/workspace \
-w /workspace \
-v ${PWD}/results:/results \
nvcr.io/nvidia/nemo:24.05 bash
Once inside the container, you can execute additional steps from a Jupyter Notebook environment.
Step 3: Download and preprocess the customization dataset
PubMedQA is a dataset for medical domain question-answering. To download the dataset, clone the pubmedqa GitHub repo, which includes steps to split the dataset into train/val/test sets.
A raw example is provided below:
"18251357": {
"QUESTION": "Does histologic chorioamnionitis correspond to clinical chorioamnionitis?",
"CONTEXTS": [ "To evaluate the degree to which histologic chorioamnionitis, a frequent finding in placentas submitted for histopathologic evaluation, correlates with clinical indicators of infection in the mother.", "A retrospective review was performed on 52 cases with a histologic diagnosis of acute chorioamnionitis from 2,051 deliveries at University Hospital, Newark, from January 2003 to July 2003. Third-trimester placentas without histologic chorioamnionitis (n = 52) served as controls. Cases and controls were selected sequentially. Maternal medical records were reviewed for indicators of maternal infection.", "Histologic chorioamnionitis was significantly associated with the usage of antibiotics (p = 0.0095) and a higher mean white blood cell count (p = 0.018). The presence of 1 or more clinical indicators was significantly associated with the presence of histologic chorioamnionitis (p = 0.019)." ],
"reasoning_required_pred": "yes",
"reasoning_free_pred": "yes",
"final_decision": "yes",
"LONG_ANSWER": "Histologic chorioamnionitis is a reliable indicator of infection whether or not it is clinically apparent." },
Given the question and the context, the goal for this tutorial is to fine-tune Llama 3 8B to give a yes/no response.
To fine-tune, convert the data to the .jsonl format, where each line is an individual example in the form of a JSON dict with the input: and output: keys for supervised learning. After preprocessing, the example looks like the following:
{
"input": "OBJECTIVE: To evaluate the degree to which histologic chorioamnionitis, a frequent finding in placentas submitted for histopathologic evaluation, correlates with clinical indicators of infection in the mother ... \nQUESTION: Does histologic chorioamnionitis correspond to clinical chorioamnionitis?\n ### ANSWER (yes|no|maybe): ",
"output": "<<< yes >>>"}
Note that the input includes the context followed by the question.
In the output, the inclusion of “<<<” and “>>>” markers enables verification of the LoRA-tuned model because the base model can produce “yes” / “no” responses based on zero-shot templates as well.
For end-to-end instructions for preprocessing, see the Jupyter Notebook tutorial.
Step 4: Fine-tune the model with NeMo framework
NeMo framework includes a high-level Python script for fine-tuning megatron_gpt_finetuning.py that can abstract away some of the lower-level API calls.
MODEL="/workspace/llama-3-8b-instruct-nemo_v1.0/8b_instruct_nemo_bf16.nemoo"
TRAIN_DS="[./pubmedqa/data/pubmedqa_train.jsonl]"
VALID_DS="[./pubmedqa/data/pubmedqa_val.jsonl]"
TEST_DS="[./pubmedqa/data/pubmedqa_test.jsonl]"
TEST_NAMES="[pubmedqa]"
# Tensor and Pipeline model parallelism
TP_SIZE=1
PP_SIZE=1
# Save results and checkpoints in this directory
OUTPUT_DIR="./results/Meta-Llama-3-8B-Instruct"
torchrun --nproc_per_node=1 \ /opt/NeMo/examples/nlp/language_modeling/tuning/megatron_gpt_finetuning.py \
exp_manager.exp_dir=${OUTPUT_DIR} \
exp_manager.explicit_log_dir=${OUTPUT_DIR} \
trainer.devices=1 \
trainer.num_nodes=1 \
trainer.precision=bf16-mixed \
trainer.val_check_interval=20 \
trainer.max_epochs=10 \
model.megatron_amp_O2=False \
++model.mcore_gpt=True \
model.tensor_model_parallel_size=1 \
model.pipeline_model_parallel_size=1 \
model.micro_batch_size=1 \
model.global_batch_size=8 \
model.restore_from_path=${MODEL} \
model.data.train_ds.num_workers=0 \
model.data.validation_ds.num_workers=0 \
model.data.train_ds.file_names=${TRAIN_DS} \
model.data.train_ds.concat_sampling_probabilities=[1.0] \
model.data.validation_ds.file_names=${VALID_DS} \
model.peft.peft_scheme="lora"
This will create a LoRA adapter in .nemo format in $OUTPUT_DIR/checkpoints.
The model.peft.peft_scheme parameter determines the technique being used. This tutorial uses LoRA, but NeMo framework supports other techniques as well, such as p-tuning, adapters, and IA3.
Training a Llama 3 70B model involves the same process, with the only differences being more memory and compute requirements, and the model to be sharded across the multiple GPUs. The recommended configuration is eight NVIDIA A100 or NVIDIA H100 80 GB GPUs, and 8-way Tensor Parallellism (TP=8, PP=1).
You can override many such configurations while running the script. For a full set of possible configurations, see the config yaml.
Step 5: Prepare your LoRA model store
Now that you have the .nemo LoRA model, it’s time to deploy it. NIM can deploy multiple LoRA adapters over the same base model. It needs a specific directory structure it can understand.
The example below shows how this “model store” should be prepared. Each LoRA adapter should be placed in a folder, the name of which will serve as a reference for dispatching requests to at inference time.
</path/to/LoRA-model-store>
├── llama3-8b-pubmed-qa
│ └── megatron_gpt_peft_lora_tuning.nemo
├── llama3-8b-lora_model_2_nemo
│ └── llama3-8b-instruct-lora_model_2.nemo
└── llama3-8b-lora_model_3_hf
├── adapter_config.json
└── adapter_model.safetensors
For this tutorial, one LoRA adapter was trained on PubMedQA, so go ahead and place that in its own directory inside a model store folder. If you have other adapters, you can replicate this process with those for a multi-LoRA deployment. Note that NVIDIA NIM supports adapters trained with the NeMo framework as well as Hugging Face PEFT.
Step 6: Deploy with NIM
After the model store is organized, deployment takes just one Docker command:
export NGC_API_KEY=<YOUR_NGC_API_KEY>
export LOCAL_PEFT_DIRECTORY=</path/to/LoRA-model-store>
chmod -R 777 $LOCAL_PEFT_DIRECTORY
export NIM_PEFT_SOURCE=/home/nvs/loras # Path to LoRA models internal to the container
export NIM_PEFT_REFRESH_INTERVAL=3600 # (in seconds) will check NIM_PEFT_SOURCE for newly added models every hour in this interval
export CONTAINER_NAME=meta-llama3-8b-instruct
export NIM_CACHE_PATH=</path/to/NIM-model-store-cache> # Model artifacts (in container) are cached in this directory
mkdir -p $NIM_CACHE_PATH
chmod -R 777 $NIM_CACHE_PATH
docker run -it --rm --name=$CONTAINER_NAME \
--runtime=nvidia \
--gpus all \
--shm-size=16GB \
-e NGC_API_KEY \
-e NIM_PEFT_SOURCE \
-e NIM_PEFT_REFRESH_INTERVAL \
-v $NIM_CACHE_PATH:/opt/nim/.cache \
-v $LOCAL_PEFT_DIRECTORY:$NIM_PEFT_SOURCE \
-p 8000:8000 \
nvcr.io/nim/meta/llama3-8b-instruct:1.0.0
The first time you run the command, the NVIDIA TensorRT-LLM-optimized Llama 3 engine will be downloaded and cached in $NIM_CACHE_PATH. This will make subsequent deployments even faster. There are several other options to configure NIM further. You can find a full list in the NIM configuration documentation.
Running this command should start a server at port 8000. Now, you’re all set to start sending inference requests.
Step 7: Sending an inference request
To create a completion, you can send a POST request to the /completions endpoint. To follow along, create a Python script in a separate terminal or start a Jupyter Notebook. The following commands use the Python requests library.
import requests
import json
url = 'http://0.0.0.0:8000/v1/completions'
headers = {
'accept': 'application/json',
'Content-Type': 'application/json'
}
# Example from the PubMedQA test set
prompt="BACKGROUND: Sublingual varices have earlier been related to ageing, smoking and cardiovascular disease. The aim of this study was to investigate whether sublingual varices are related to presence of hypertension.\nMETHODS: In an observational clinical study among 431 dental patients tongue status and blood pressure were documented. Digital photographs of the lateral borders of the tongue for grading of sublingual varices were taken, and blood pressure was measured. Those patients without previous diagnosis of hypertension and with a noted blood pressure \u2265 140 mmHg and/or \u2265 90 mmHg at the dental clinic performed complementary home blood pressure during one week. Those with an average home blood pressure \u2265 135 mmHg and/or \u2265 85 mmHg were referred to the primary health care centre, where three office blood pressure measurements were taken with one week intervals. Two independent blinded observers studied the photographs of the tongues. Each photograph was graded as none/few (grade 0) or medium/severe (grade 1) presence of sublingual varices. Pearson's Chi-square test, Student's t-test, and multiple regression analysis were applied. Power calculation stipulated a study population of 323 patients.\nRESULTS: An association between sublingual varices and hypertension was found (OR = 2.25, p<0.002). Mean systolic blood pressure was 123 and 132 mmHg in patients with grade 0 and grade 1 sublingual varices, respectively (p<0.0001, CI 95 %). Mean diastolic blood pressure was 80 and 83 mmHg in patients with grade 0 and grade 1 sublingual varices, respectively (p<0.005, CI 95 %). Sublingual varices indicate hypertension with a positive predictive value of 0.5 and a negative predictive value of 0.80.\nQUESTION: Is there a connection between sublingual varices and hypertension?\n ### ANSWER (yes|no|maybe): "
data = {
"model": "llama3-8b-pubmed-qa",
"prompt": prompt,
"max_tokens": 128
}
response = requests.post(url, headers=headers, json=data)
response_data = response.json()
print(json.dumps(response_data, indent=4))
The output looks like the following:
{
"id": "cmpl-403d22baa7c3470eb468ee8a38033e1f",
"object": "text_completion",
"created": 1717493046,
"model": "llama3-8b-pubmed-qa",
"choices": [
{
"index": 0,
"text": " <<< yes >>>",
"logprobs": null,
"finish_reason": "stop",
"stop_reason": null
}
],
"usage": {
"prompt_tokens": 412,
"total_tokens": 415,
"completion_tokens": 3
}
}
This example returned a text output "<<< yes >>>” along with other metadata. If you recall from a few steps ago, this is the format it was trained on. Running inference over the whole PubMedQA test set and calculating accuracy gives the following metrics:
Accuracy 0.786000
Macro-F1 0.584112
Summary
Voila! You have successfully customized a Llama 3 8B Instruct model and deployed it using NVIDIA NIM. With a few training steps and a short training time, you can achieve a reasonably accurate model, when compared to the PubMedQA Leaderboard. The full tutorial also includes instructions on calculating these metrics. It’s possible to further tune the hyperparameters to gain better accuracy, and the state-of-the-art training optimizations with NeMo framework enable faster iterations.
To further simplify generative AI customization, the NeMo team has announced an early access program for NVIDIA NeMo Customizer microservice. This high-performance, scalable service simplifies fine-tuning and aligning LLMs for domain-specific use cases. Leveraging well-known microservices and API architecture, it helps enterprises bring solutions to market faster. Apply for early access.